Introduction
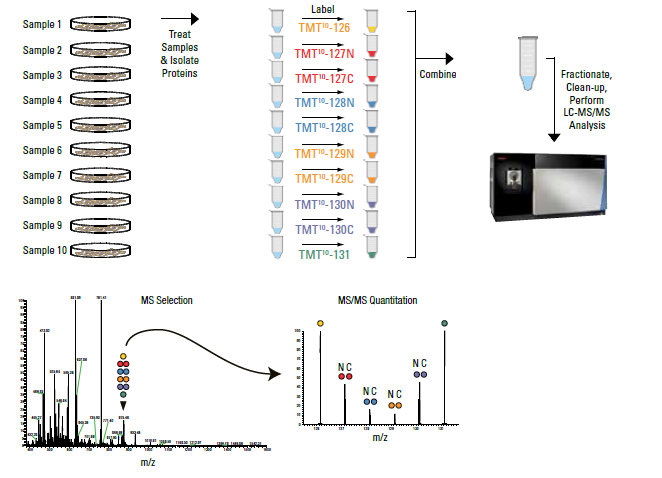
Tandem Mass Tag (TMT) technology is a powerful technique for comparing protein (and peptide) amounts between six to eighteen conditions. This technique involves labeling samples after digestion, meaning this technique can be used on in vivo samples, unlike SILAC.
TMT or TMTpro samples are to be submitted in solution or on-bead, not in gel. It is the responsibility of the customer to ensure that similar amounts of protein is present in each sample and that the samples are contaminant free. A full description of TMT is outside the scope of this website, but details can be found at:
https://en.wikipedia.org/wiki/Tandem_mass_tag
https://www.thermofisher.com/us/en/home/life-science/protein-biology/protein-mass-spectrometry-analysis/protein-quantitation-mass-spectrometry/tandem-mass-tag-systems.html
Samples to be analyzed by TMT quantitation should be submitted either on-bead (if applicable), or as in-solution samples in suitable buffer (RIPA buffer or 50 mM triethylammonium bicarbonate and 5% SDS. Other buffers may be suitable, please contact us with questions). Analysis of acquired mass-spectrometry data will be performed with Proteome Discoverer 3.0 software.
What Does it Cost?
$1530 for TMT 6-plex, $2040 for TMT 10-plex, $3672 for TMTpro 18-plex. Any number of samples between 2-18 can be sumbitted in a single batch, and prices will be adjusted for labels that are not used. High-pH fractionation is an additional $1,020 per set of up to 18 samples and can significantly increase the number of proteins identified in complex samples.
What We Do
We run your TMT/TMTpro samples on our Orbitrap Eclipse mass spectrometer platform, using an appropriate LC-MS/MS method including real-time search. We identify and quantify peptides and proteins from samples using Proteome Discoverer 3.0 and send you a spreadsheet including a list of proteins present and the relative amount of each protein in each sample.
What You Must Do
Prior to submitting TMT/TMTpro samples you should:
- Contact us before beginning the experiment to discuss experimental design. Ensure all samples have similar amounts of protein.
- Put each sample into an Eppendorf 1.5ml tube, which has been rinsed with 50% organic solvent and millipure water.
- Follow the general guidelines for sample submission.
Experimental Design
Appropriate experimental design and statistical analysis of results are key to a successful quantitative proteomics experiment. You must contact us before beginning your quantitative experiment. Biological and technical repeats are necessary for robust results, so quantitative experiments can be lengthy and expensive. Careful consideration upfront prevents wasted time and money later.
Any Questions?
If you are planning TMT or TMTpro samples please always contact ProteomicsCore@UTSouthwestern.edu before you prepare and submit your samples.