 Introduction
Introduction
Modern LC-MS/MS platforms can identify several thousands of proteins from a single sample. We realize that many different types of samples can be submitted for proteomics analsyis, so we accept a wide range of sample formats, including:
- On-bead (magnetic beads preferred)
- In-solution
- SDS-PAGE gel
Our instrumentation allows the characterization of complex samples without the need to resolve on a gel and excise bands. Comparison between complex samples is often better performed by complex mixture analysis than by running a gel and cutting out visibly different bands. The mass-spectrometer will observe more differences than are visible with Coomassie or silver staining.
What Does it Cost?
$204 per sample or 10 mm slice of gel (90 min HPLC gradient) or $306 for a 3hr HPLC gradient.
What We Do
We run your samples on our QExactive HF or Orbitrap Fusion Lumos mass spectrometers, using a 90 minute or 3 hour reverse-phase LC-MS/MS method. We identify proteins from samples using Proteome Discoverer 3.0 and e-mail an Excel summary of identifications. We can provide a single report comparing the content of multiple samples, including label-free quantitation (at no extra charge).
What You Must Do (bead/solution samples)
Samples should be submitted in 1.5 mL Eppendorf tubes. Details regarding buffer composition (if applicable) and protein concentration or amount must be included.
What You Must Do (gel samples)
To submit a complex mixture gel sample you must:
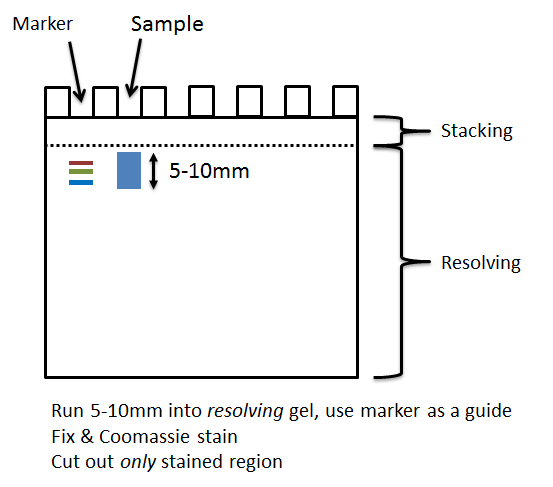
- Run your sample ~1cm into the resolving part of a pre-cast SDS PAGE gel. Your sample should proceed into the resolving area, and not remain in any stacking area of the gel.
- Stain your gel with Coomassie Blue – all complex mixtures must be stained. If you use a stain other than Coomassie Blue you must include an acetic acid protein fixation step.
- Cut out the stained area as one or more 5-10cm slices. Each slice will be a separate sample, analyzed and charged. Do not cut out the very top of the gel (loading/stacking) area, which is likely to have detergent from your buffer on its surface, and try to avoid including empty gel around the protein.
- Adhere to our guidance when cutting out the band, ensuring you dice each band into 1mm cubes (no smaller) etc. and ensure your slice(s) are no larger than 10mm of gel.
- Place each slice into an Eppendorf 1.5ml tube, which has been rinsed with 50% organic solvent (MeOH or ACN) and milliQ/ultrapure water.
- Follow the general guidelines for sample submission.

We offer free label-free quantitative comparison for complex mixtures. It is useful to identify larger differences in abundance of proteins between samples. Accuracy depends on the complexity of the sample, and abundance of the proteins quantified. It is not suitable to accurately quantify relatively small fold-changes, e.g. 2-fold or less (dependent on sample).
Contact us before submission if you would like a quantitative analysis. Depending on your experiment it is likely that biological and technical replicates will be required for a useful outcome.
The Orbitrap Fusion Lumos instrument currently used for most service work can identify over 5000 proteins per run from an abundant whole lysate. However, depth of coverage depends on the the amount of protein present, and not exceeding the maximum capability of the mass-spectrometer. If your sample is likely to contain >3000 proteins you should consider using the Lumos for analysis. Contact us if you have any doubts about which service to choose.
Running a silver-stain gel using a small amount of the sample can be useful to test sample quality and to determine the complexity of the sample.
Warnings & Limitations
Complex mixture analysis on our instrumentation is powerful and very cost-effective. However, it is important to match the complexity of your sample to an appropriate run-time and appropriate mass spectrometer. Consider speaking to us before submitting your first sample, or if you have any doubts about the complexity of your sample.
We can run searches for post-translational modifications against complex mixtures, but are not able to review and validate results due to constraints on staff time – this is left to the customer. We provide a more comprehensive PTM service for purified protein samples or for samples enriched for a sepcific PTM.
Any Questions?
If you have any questions please contact ProteomicsCore@UTSouthwestern.edu before you prepare and submit your samples.