Introduction
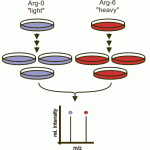 Stable Isotope Labeling by Amino Acids in Cell Culture (SILAC) is a powerful technique for comparing protein (and peptide) amounts between two or three conditions. Developed in the lab of Matthias Mann it is one of the most widely used labeled quantitation methods in proteomics.
Stable Isotope Labeling by Amino Acids in Cell Culture (SILAC) is a powerful technique for comparing protein (and peptide) amounts between two or three conditions. Developed in the lab of Matthias Mann it is one of the most widely used labeled quantitation methods in proteomics.
In SILAC experiments cells for each condition are grown in media that contains different stable isotopes of amino acids such that an identical peptide from each condition has a slightly different mass. The peptides are visible in the mass-spectrometer as doublets or triplets, separated by the mass of the isotope label on the amino acids they contain. Typically SILAC uses heavy labeled Arg and Lys, since every tryptic peptide will contain at least one Arg or Lys, and produce a doublet or triple for quantitation.
Kits containing SILAC media can be purchased from commercial vendors:
https://www.thermofisher.com/order/catalog/product/A33972
or labelled amino acids may be purchased separately to create SILAC media in the lab.
The Proteomics Core can accept SILAC labelled samples for protein identification and quantitation. Analysis of acquired mass-spectrometry data will be performed with either Proteome Discoverer or the MaxQuant software from the Mann Lab.
What Does it Cost?
$224 per 5-10mm gel slice. Note you will need to budget for the cost of preliminary experiments (see below).
What We Do
We run your SILAC samples on our Orbitrap Fusion Lumos mass-spectrometry platforms, using an appropriate LC-MS/MS method. We identify and quantify peptides and proteins from samples using Proteome Discoverer 2.4 or the MaxQuant software from the Mann Lab. We provide an Excel summary of identifications and the SILAC quantitation.
What You Must Do
To submit a SILAC sample you should:
- Contact us before beginning the experiment to discuss experimental design, SILAC labeling, and steps needed to check label incorporation, mixing etc. (See below).
- Grow sufficient cells for the experiment you are planning. We recommend one or more 10cm dishes for whole lysate experiments, and 5 x 15cm dishes for IP-type experiments (see below).
- Perform SILAC labelling of your cell lines, treat as required, and mix into a single light/heavy sample.
- Run your SILAC labelled protein mixture into the top of the resolving portion of an SDS-PAGE gel. Run in 5-10mm to submit as a single sample. Run in further to submit as multiple 5mm slices for increased coverage.
- Cut out the stained area as one or more 5-10mm slices. Each slice will be a separate sample, analyzed and charged. Do not cut out the very top of the gel (loading/stacking) area, which is likely to have detergent from your buffer on it’s surface.
- Adhere to our guidance when cutting out the band, ensuring you dice each band into 1mm cubes (no smaller) etc. and ensure your slice(s) are no larger than 10mm of gel.
- Place each slice into an Eppendorf 1.5ml tube, which has been rinsed with 50% organic solvent and millipure water.
- Follow the general guidelines for sample submission.
We strongly prefer that you label your heavy cells with Arg and Lys labels, that have different mass shifts (Lys8, Arg10 preferred). This supplies additional information in the analysis step – we know if a peptide contains Arg/Lys from the observed mass shift. SILAC labeling kits are available from various sources. Note that many only contain the Lys label in the kit – you must order the Arg label separately. Bulk purchase of media and labelled amino acids may be cost effective if you are planning a series of SILAC experiments.
Experimental Design
Appropriate experimental design and statistical analysis of results are key to a successful quantitative proteomics experiment. You must contact us before beginning your quantitative experiment. Biological and technical repeats are necessary for robust results, so quantitative experiments can be lengthy and expensive. Careful consideration upfront prevents wasted time and money later.
Preliminary QC Experiments
It is vital to perform preliminary QC experiments prior to the main SILAC experiment to ensure the best possibility of success:
1) Label Incorporation – When growing a new cell line in heavy medium for the first time you must submit a QC sample of whole lysate, from a 6cm dish of cells, to check label incorporation. Incomplete incorporation of the SILAC label into proteins will affect the dynamic range and accuracy of the analysis.
2) Mixing Ratio – When performing SILAC experiments it is important that you can accurately mix 1:1 amounts of light and heavy material. After the incorporation test you must submit a 1:1 mix of untreated whole-cell lysates for quantification. We advise if your 1:1 mixing is accurate. Inaccurate mixing affects the dynamic range and accuracy of the analysis.
3) Purification Test – If you are performing a SILAC experiment that involves a purification step you should submit an unlabeled test sample for the purification step. This will let us advise you on whether your purification brings down enough material, if there is too much background etc. This step can save the expense of troubleshooting your purification while growing cells in expensive SILAC medium.
The Real SILAC Experiment
After preliminary tests you will be ready to perform your actual SILAC experiment, with differential treatment of the light and heavy cells. In general we suggest that you start from the following material:
For experiments studying whole cell lysate – at least 1 x 10cm dish of cells for light and heavy. We recommend using more starting material and fractionating the sample to get good coverage in a lysate experiment. Contact us.
For IP-type experiments – 5 x 15cm dishes of cells for heavy and light. You may be purifying an over-expressed protein, but you will usually be interested in what comes down with it. These co-purified proteins will be at endogenous levels, which may be very low. A large amount of starting material is necessary to look deep into the proteome.
Any Questions?
If you are planning a SILAC experiment please always contact ProteomicsCore@UTSouthwestern.edu before you prepare and submit your samples.